
notepad++) they look like: And I want dot format of identities like and then save it in txt file. I need some way to get the first 10 bases (and then I was planning on doing it again for the last 10 bases). I have fasta file with alingned several sequences (from MUSCLE), when I open it (e.g.

So here's an example: >gi|2765658|emb|Z78533.1|CIZ78533 C.irapeanum 5.8S rRNA gene and ITS1 and ITS2 DNAĬGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTGATGAGACCGTGGAATAAACGATCGAGTGĪATCCGGAGGACCGGTGTACTCAGCTCACCGGGGGCATTGCTCCCGTGGTGACCCTGATTTGTTGTTGGG A FASTQ file usually uses four lines per sequence. It is mainly used for storing the output of high-throughput sequencing instruments. The transcripts.txt file contains the list transcripts IDs that I want to export (both the IDs and the sequences) from assembly.fasta to selectedtranscripts.fasta. This functions as a placeholder until GenBank assigns. I want to extract specific fasta sequences from a big fasta file using the following script, but the output is empty. 2) Create a short, unique sequence ID (SeqID) that you can use for each sequence. If you use a word processing program, you must save the file as plain ASCII text in order to retain the FASTA format.
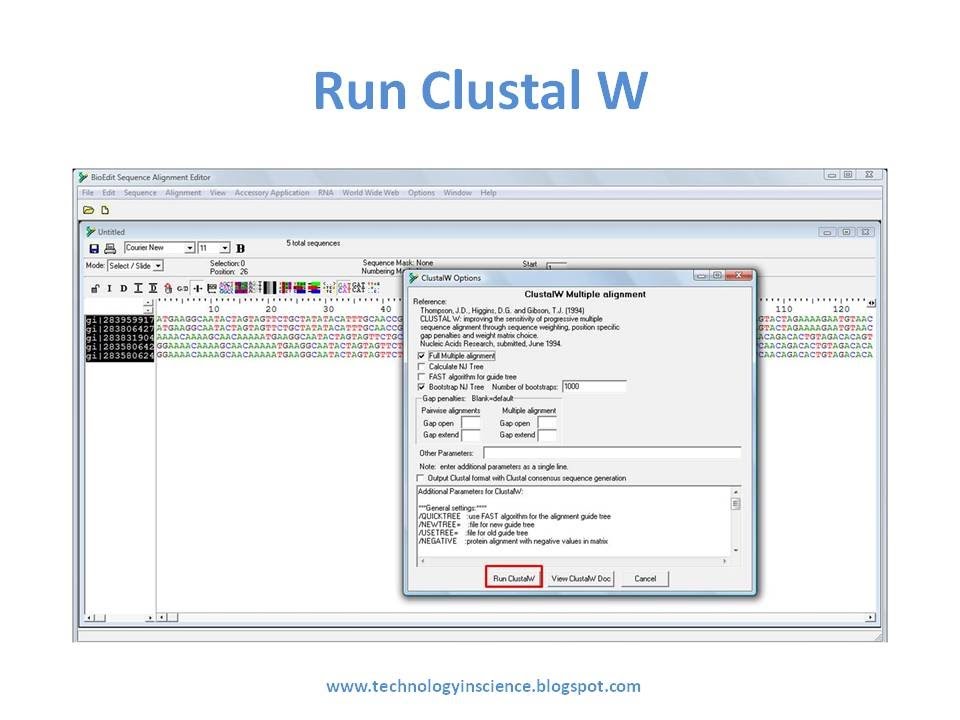
BIOEDIT EXPORT TXT TO FASTA FORMAT HOW TO
FASTQ is a text-based format for storing both a biological sequence (usually nucleotide sequence) and its corresponding quality scores. Here is how to create the FASTA file: 1) We strongly recommend that you use a text editor. Worst comes to worst, I could just use the bases if there's no way to keep the sequence info. A sequence file in FASTQ format can contain several sequences. I need to get the first 10 bases from each sequence and put them in one file, preserving the sequence info from the FASTA format. Ok so I need to extract part of a sequence from a FASTA file, using python (biopython, )


 0 kommentar(er)
0 kommentar(er)
